Parameter estimates for regression: least squares, gradient descent and monte carlo methods22/8/2015
[data and R code for this post are available on github]
In this post I will cover three ways to estimate parameters for regression models; least squares, gradient descent and Monte Carlo methods. The aim is to introduce important methods widely used in machine learning, such as gradient descent and Monte Carlo, by linking them to a common "use case" in the different data science communities, such as linear regression. Regression is one of the first prediction methods invented, and the three approaches I will be discussing, are typically used by three different communities. Least squares is probably the most common method, mainly employed by frequentist statisticians and also used as the default method for many easy to use packages in R or Python (e.g. the lm function in R). Gradient descent is the machine learning approach to the problem. My favorite resource on the topic is the famous machine learning course by Andrew Ng on Coursera. In machine learning, the overall approach to problem solving and prediction is rather different compared to more classical statistics, even though it heavily relies on statistics (check out this answer by Sebastian Raschka on the origins of machine learning, I think it makes my point clear). I think one of the reasons why Andrew Ng uses gradient descent in the first place, instead of other methods like the least squares, is probably that he wants to stress the importance of the method in the machine learning community. By using gradient descent, he highlights how in machine learning it's often more important to approximate a solution by defining an iterative procedure able to efficiently explore the parameter space, instead of obtaining an exact analytical solution. Finally, there is the Bayesian way of doing things. Monte Carlo methods are powerful tools to explore the parameter's space and obtain the full posterior distribution, instead of just point estimates. This is something that requires a bit more introduction and explanation, and while I tried to do so in the remaining of this blog post, this is certainly far from being a comprehensive resource on Bayesian modeling. For the ones that want to dig deeper into regression and Bayesian approaches I would suggest reading Gelman's book on hierarchical modeling, one of my favorite resources in the field.
4 Comments
[For this analysis I used the term/preterm dataset that you can find on Physionet. My data and code are also available on github]
A couple of weeks ago I read this post about cross-validation done wrong. During cross-validation, we are typically trying to understand how well our model can generalize, and how well it can predict our outcome of interest on unseen samples. The author of the blog post makes some good points, especially about feature selection. It is indeed common malpractice to perform feature selection before we go into cross-validation, something that should however be done during cross-validation, so that the selected features are only derived from training data, and not from pooled training and validation data. However, the article doesn’t touch a problem that is a major issue in most clinical research, i.e. how to properly cross-validate when we have imbalanced data. As a matter of fact, in the context of many medical applications, we have datasets where we have two classes for the main outcome; normal samples and relevant samples. For example in a cancer detection application we might have a small percentages of patients with cancer (relevant samples) while the majority of samples might be healthy individuals. Outside of the medical space, this is true (even more) for the case for example of fraud detection, where the rate of relevant samples (i.e. frauds) to normal samples might be even in the order of 1 to 100 000. problem at hand
The main motivation behind the need to preprocess imbalanced data before we feed them into a classifier is that typically classifiers are more sensitive to detecting the majority class and less sensitive to the minority class. Thus, if we don't take care of the issue, the classification output will be biased, in many cases resulting in always predicting the majority class. Many methods have been proposed in the past few years to deal with imbalanced data. This is not really my area of research, however since I started working on preterm birth prediction, I had to deal with the problem more often. Preterm birth refers to pregnancies shorter than 37 weeks, and results in about 6-7% of all deliveries in most European countries, and 11% of all deliveries in the U.S., therefore the data are quite imbalanced.
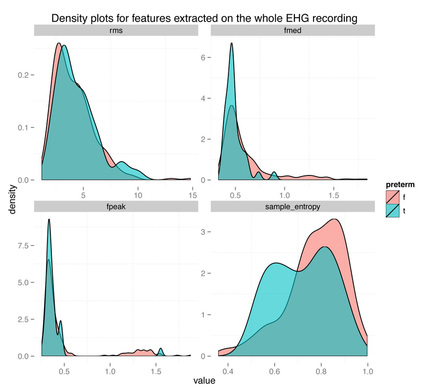
I recently came across two papers [1, 2] predicting term and preterm deliveries using Electrohysterography (EHG) data. The authors used one single cross-sectional EHG recording (capturing the electrical activity of the uterus) and claimed near perfect accuracy in discriminating between the two classes (AUC value of 0.99 [2], compared to AUC = 0.52-0.60 without oversampling). This seemed to me like a clear case of overfitting and bad cross-validation, for a couple of reasons. First of all, let’s just look at the data:
The density plots above show the feature's distribution for four features over the two classes, term and preterm (f = false, the delivery was not preterm, in light red, t = true, the delivery was preterm, in light blue). As we can see there is really not much discriminative power here between conditions. The extracted features are completely overlapping between the two classes and we might have a "garbage in, garbage out" issue, more than a "this is not enough data" issue.
Just thinking about the problem domain, should also raise some doubts, when we see results as high as auc = 0.99. The term/preterm distinction is almost arbitrary, set to 37 weeks of pregnancy. If you deliver at 36 weeks and 6 days, you are labeled preterm. On the other hand, if you deliver at 37 weeks and 1 day, you are labeled term. Obviously, there is no actual difference due to being term or preterm between two people that deliver that close, it's just a convention, and as such, prediction results will always be affected and most likely very inaccurate around the 37 weeks threshold. Since the dataset used is available for anyone to download and use from Physionet, in this post I will partially replicate the published results, and show how to properly cross-validate when oversampling data. Maybe some clarification on this issue will help in avoiding the same mistakes in the future. |
Marco ALtiniFounder of HRV4Training, Advisor @Oura , Guest Lecturer @VUamsterdam , Editor @ieeepervasive. PhD Data Science, 2x MSc: Sport Science, Computer Science Engineering. Runner Archives
May 2023
|
 RSS Feed
RSS Feed